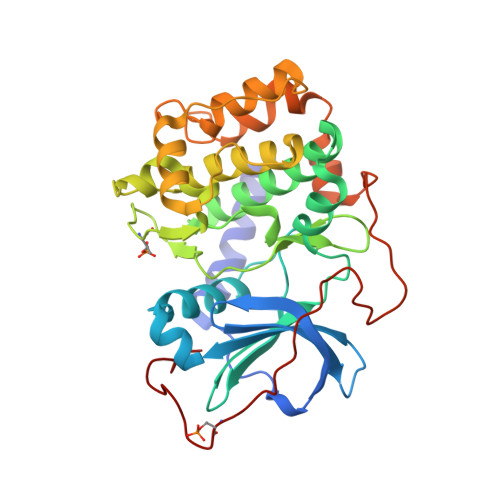
Discovery of 2-pyrimidyl-5-amidothiophenes as potent inhibitors for AKT: synthesis and SAR studies
Lin, X., Murray, J.M., Rico, A.C., Wang, M.X., Chu, D.T., Zhou, Y., Del Rosario, M., Kaufman, S., Ma, S., Fang, E., Crawford, K., Jefferson, A.B.(2006) Bioorg Med Chem Lett 16: 4163-4168
- PubMed: 16765046
- DOI: https://doi.org/10.1016/j.bmcl.2006.05.092
- Primary Citation of Related Structures:
2GU8 - PubMed Abstract:
A series of 2-pyrimidyl-5-amidothiophenes has been synthesized and evaluated for AKT inhibition. SAR studies resulted in potent inhibitors of AKT with IC(50) values as low as single digit nanomolar as represented by compound 2aa. Compound 2aa showed cellular activity including antiproliferation and downstream target modulation. Selectivity profile is described. A co-crystal of 2aa with PKA is determined and discussed.
Organizational Affiliation:
Small Molecule Drug Discovery, Biopharma Research, Chiron Corporation, Emeryville, CA, USA. xiaodong_lin@chiron.com